1. What is the FibroAtlas?
FibroAtlas database 1.0 (http://biokb.ncpsb.org/FibroAtlas/), which is an online database provided by National Center for Protein Science (Beijing); it contains a comprehensive list of fibrosis-related genes with their correspondent evidences in literature, derived by text mining and manual curation.
2. How to search the FibroAtlas, the FibroAtlas-related diseases and their supporting literature evidence?
In order to serve better, we provide three query approaches for searching FibroAtlas: search by protein name, search by either nucleotide sequence or protein sequence.
For the query by gene name, the user can input a gene name in the search box of "Gene Symbol", and see a drop-down list with auto-completed gene symbols present in the FibroAtlas. After selecting one of them and clicking the 'Search' button, the search engine will run and return the queried results containing a table showing the queried gene, related diseases and the supporting literature evidence. If users click the 'Reset' button, all current search terms will be deleted.
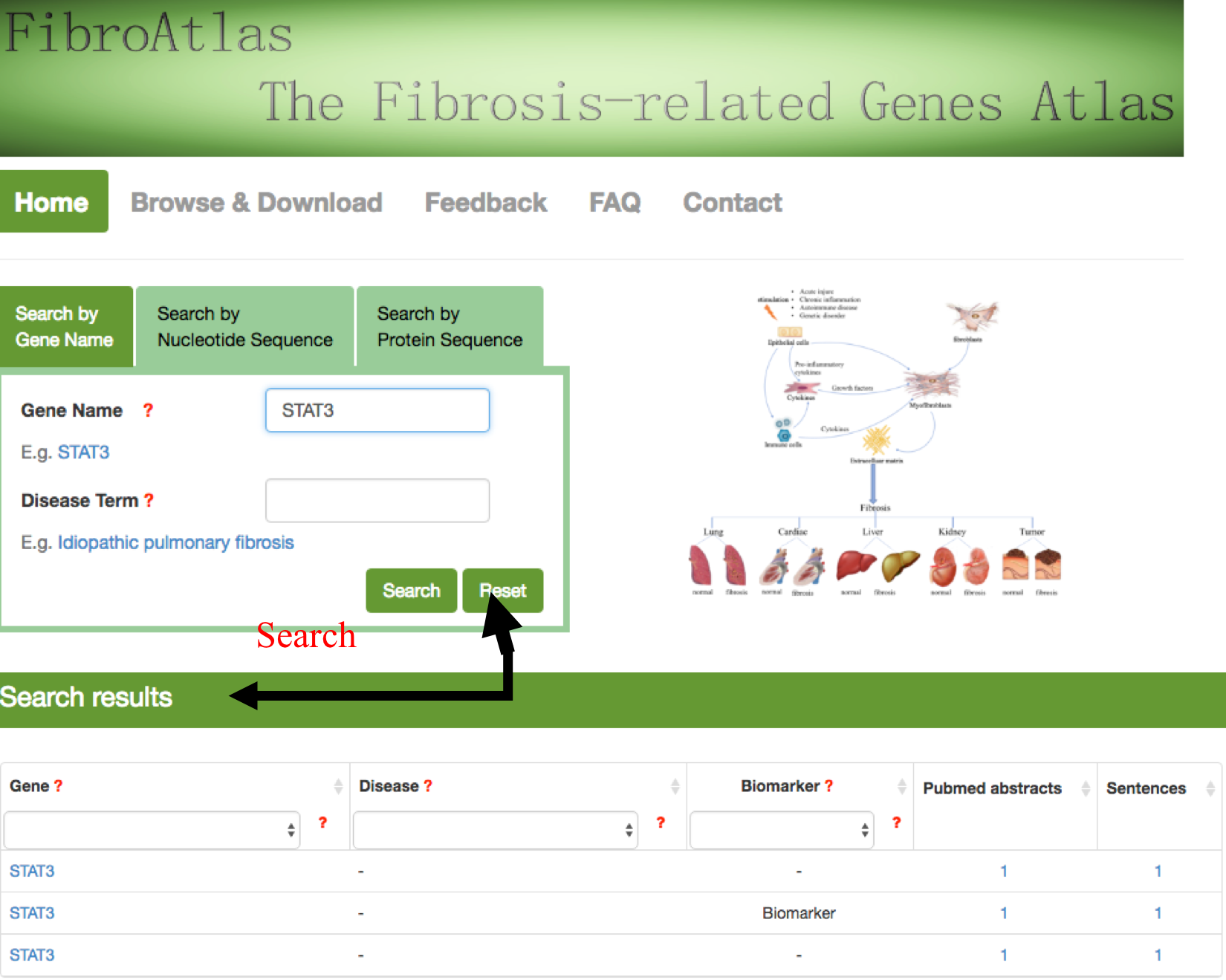
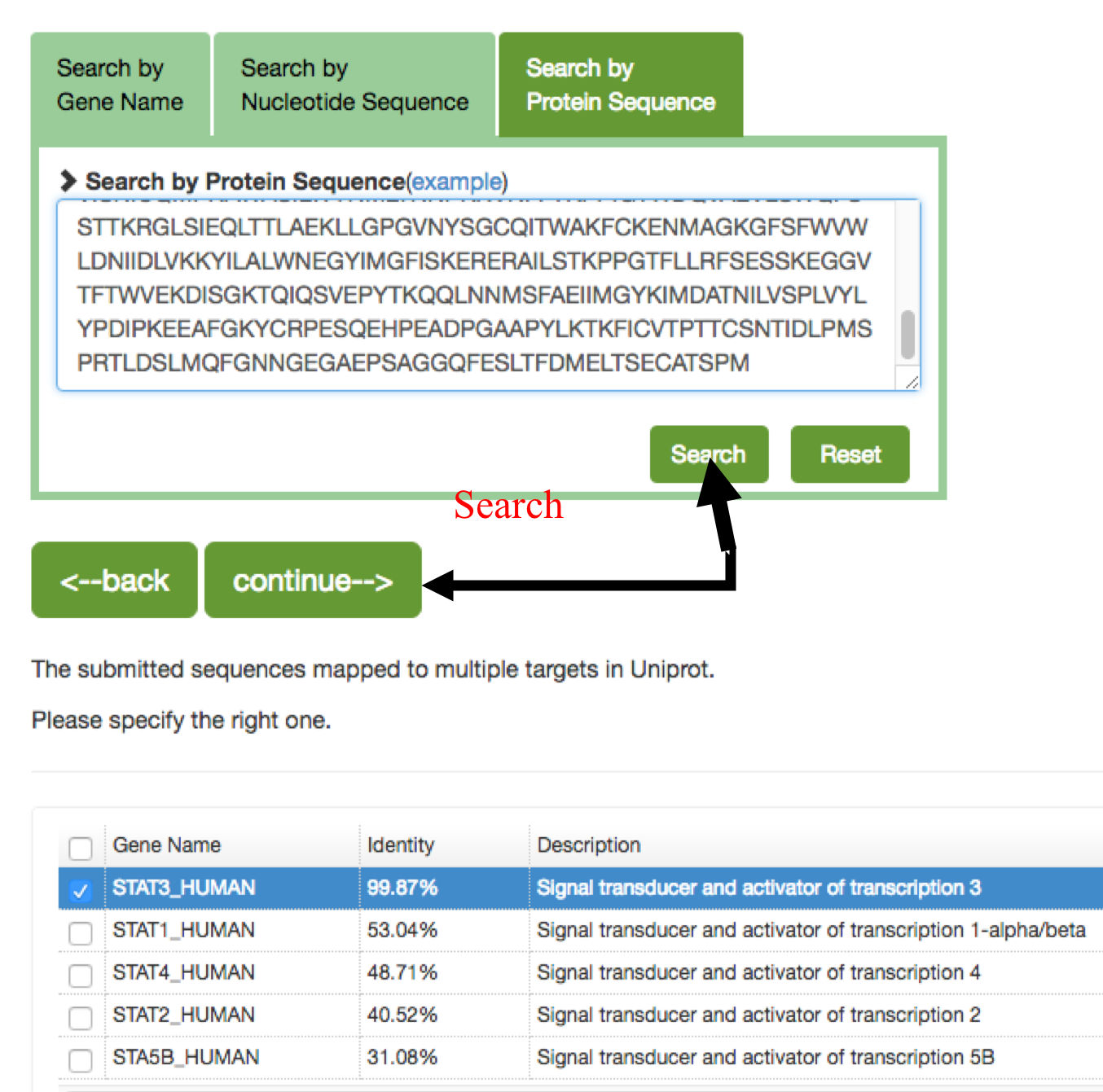
The users also can search the gene by Nucleotide sequence or protein sequence, the sequence identity score from BLAST will be listed in the parenthesis after the description. Users can specify the matched gene symbol and click “continue” for result page.
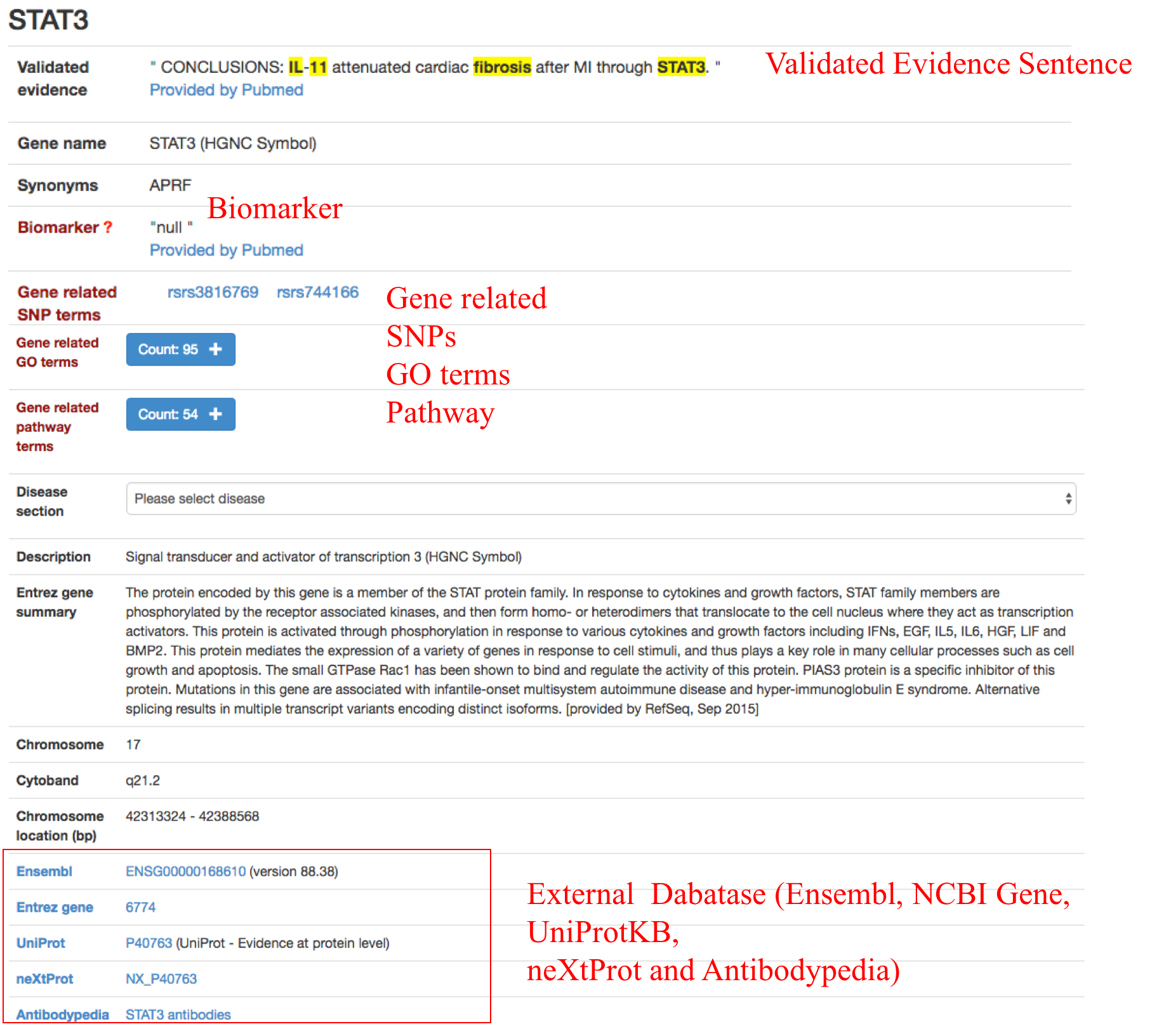
Clicking the hyperlink of the gene symbol for an individual fibrosis-related gene in the 'Gene' column, users can see well annotated information of the gene and the cross references to external databases (i.e. Ensembl, NCBI Gene, UniProtKB, neXtProt and Antibodypedia).
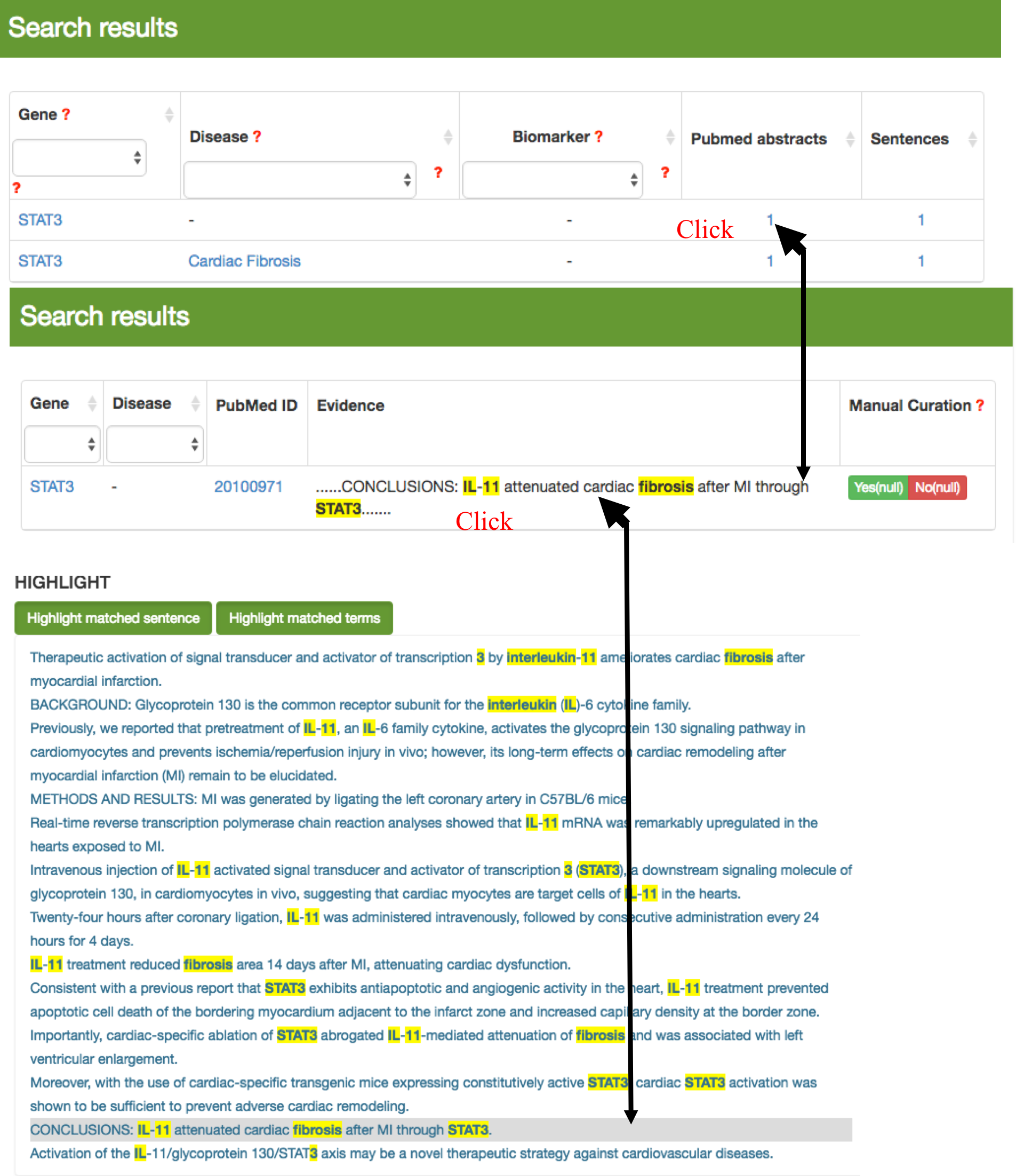
The user can also input words or select a term in the drop list in the text box to filter the listed genes or diseases from the results by sub-string match. In addition, clicking the small triangle in the head of each column in the table, the correspondent results in the table will be resorted in ascending/descending order by the column.
Clicking the number of the evidence abstracts or sentences, a table containing gene, disease, the PubMed ID, the evidence sentence and the manual validation information will be displayed to the users. The user can click on the evidence to see the original abstract with the highlighted key words, i.e. gene symbols and disease terms.
3. How to browse and download the data of FibroAtlas?
The page of "Browse & Download" shows three different methods for browsing detailed information on fibrosis-related genes: browse by gene, browse by disease and browse by biomarker. All the information for fibrosis-related genes and their supporting literature evidences can be downloaded for further analysis.
4.How does our team collect and curate the fibrosis-related genes and supporting evidence in literature to construct FibroAtlas?
To obtain a complete list of publications for fibrosis-related genes, we performed a comprehensive search for fibrosis-related literature abstracts in PubMed. Gene-nomenclature recognition and extraction from these abstracts for human fibrosis-related gene candidates were performed by self-developed ontology-based bio-entity recognizer, which has the precision, recall, F-measure of 0.810, 0.883, 0.845 against the CRAFT corpus for gene/protein recognition based on Protein Ontology (PR), and is on par with current state-of-the-art biomedical annotation systems like BeCAS.
A list of human fibrosis-related genes together with their related diseases and evidence from PubMed abstract was compiled in the following three steps.
First, PubMed abstract and sentences containing the keywords of either ‘fibrosis’ or ‘allergic’ or ‘anaphylaxis’ or ‘allergic reaction ’or ‘hypersensitivity’ or ‘atopic’ or their lexical variants were collected. And 114,973 abstracts and 227,458sentences were obtained.
Second, a list of human genes which co-occured with the fibrosis-related keywords at single-sentence level were recognized and extracted from these sentences by our bio-entity recognizer based on Protein Ontology.
Third, all 3,150 candidates from 10243 original abstracts and 13226 sentences were manually curated by our experts and 1439 genes were finally selected as human fibrosis-related genes.
Disease terms were identified from PubMed abstracts by bio-entity recognizer based on Human Disease Ontology (DO). Associations between fibrosis-related genes/proteins and human disease terms were obtained based on sentence-level co-occurrence.
Furthermore, the biomarker roles of certain genes/proteins are recognized and marked as well. All extracted fibrosis-related genes/proteins, human disease terms as well as their biomarker roles were loaded into MySQL database.
5. How to guarantee data quality in FibroAtlas?
Three rounds of strict manual curation for the candidate fibrosis-related genes have taken to guarantee data quality. First, two experienced researchers independently checked all candidate genes and supporting evidence ; second, these selected genes with corresponding evidences were submitted to an internal review, in which each of the selected genes names were manually reviewed by a reviewer team consisting of three experts; third, all co-authors were asked to randomly check fibrosis-related genes from our website to make sure that all identified genes stored in our database are high credible fibrosis-related genes. Finally, we obtained 1195 genes and 577 associated diseases. All the evidence sentences that regarded as validated evidence have been manually curated and included on the top of each fibrosis-related gene page.
6. What is meaning of "biomarker" in the database?
A biomarker is a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.
7. What is the meaning of "Validated evidence" and "Disease section"?
"Disease section" means a classification of human disease associated with allergy-related genes.All disease terms were extracted from PubMesd abstracts by our bio-entity recognizer based on Human Disease Ontology.
"Validated evidence" means that the sentence which underwent strict three rounds of manual curation was selected as "validated evidence" to represent the correlation between the gene and allergy.
8. What is the meaning of "Highlight matched sentence" or "Highlight matched terms"?
The sentences or terms matching key words of genes or key words of allergy or disease terms that based on Human Disease Ontology, are extracted from PubMesd abstracts by our bio-entity recognizer and highlighted in our database.
9. What does "Gene Name" mean?
Gene names mean formal gene symbols and their synonyms provided by RefSeq.
10. What does "Disease Term" mean?
Disease terms mean standardized disease names and their common synonyms provided by Human Disease Ontology.
11. Can the users provide their feedback to the evidence of fibrosis-related genes?
Yes, "Feedback" feature was provided to our website, with which the users can submit new genes to our database manually; the database will be updated periodically in future. we also added a manual curation function to the evidence sentences of disease terms, with which the users can provide their feedback by simply clicking the "Yes" or "No" button. In addition, the users can also send email to us for further questions or potential collaborations.
