
1、What is the AAg Atlas portal?
AAg Atlas portal is an online resource and analysis portal provided by National Center for Protein Science (Beijing); it contains a comprehensive list of curated human autoantigens (AAgs) and correspondent supporting evidence from literature and databases (OmicsDI, GEO, AAgMarker, ArrayExpress and PMD).
2、What is the difference between AAg Atlas portal and other AAg databases?
The number of AAgs in our database is 6,919 and 3,575 more AAgs than AAgMarker 1.0 database (http://bioinfo.wilmer.jhu.edu/AAgMarker/) and AAgAtlas 1.0 database (http://biokb.ncpsb.org/aagatlas/) respectively, constituting the largest AAg database to date (Figure 1-3). Furthermore, we have developed a portal (http://biokb.ncpsb.org/aagatlas_portal/) for users to easily browse and analyze the expression and biological pathways of AAg genes on our website (Figure 1).
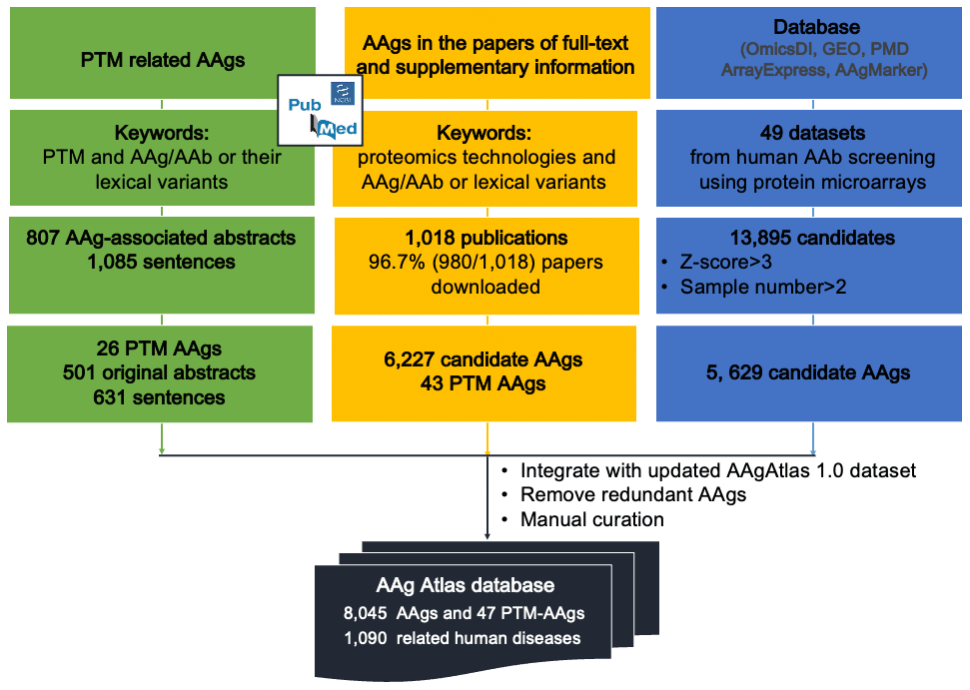
Figure 1. The workflow of human AAg Atlas portal database construction.
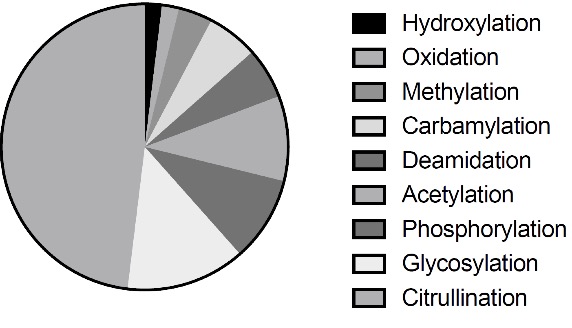
Figure 2. The post-translational modifications that are related to AAb recognition.
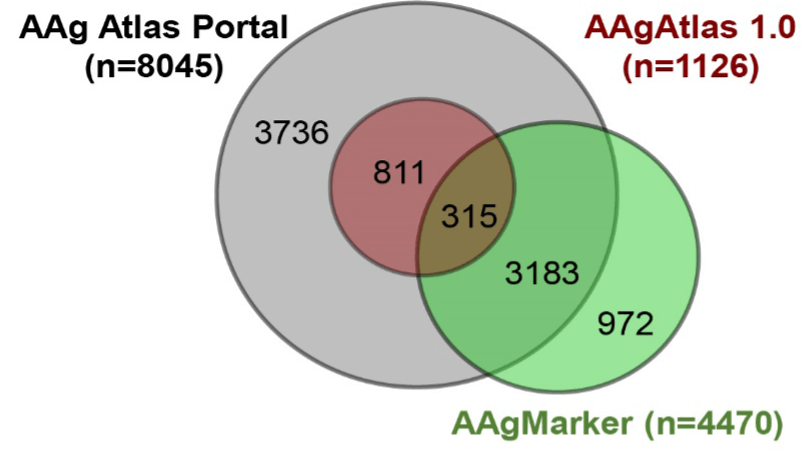
Figure 3. Comparison of human AAgs in different databases by Venn diagram analysis.
3、How to search the AAgs, the AAg-related diseases and their supporting evidence?
Two query approaches are provided for searching AAgs, query by gene symbol and query by disease term.
For the query by gene symbol, the user can enter a gene symbol in the “Gene Symbol” search box, and a drop-down menu will provide auto-completed gene symbols present in the AAgAtlas. After selecting one of them and clicking the ‘Search’ button, the search engine will run and return the queried results containing a table showing the AAg gene, related diseases as well as the supporting evidence from PubMed abstract, full-text articles or microarray dataset. If user click the ‘Reset’ button, all current search terms will be deleted (Figure 4).
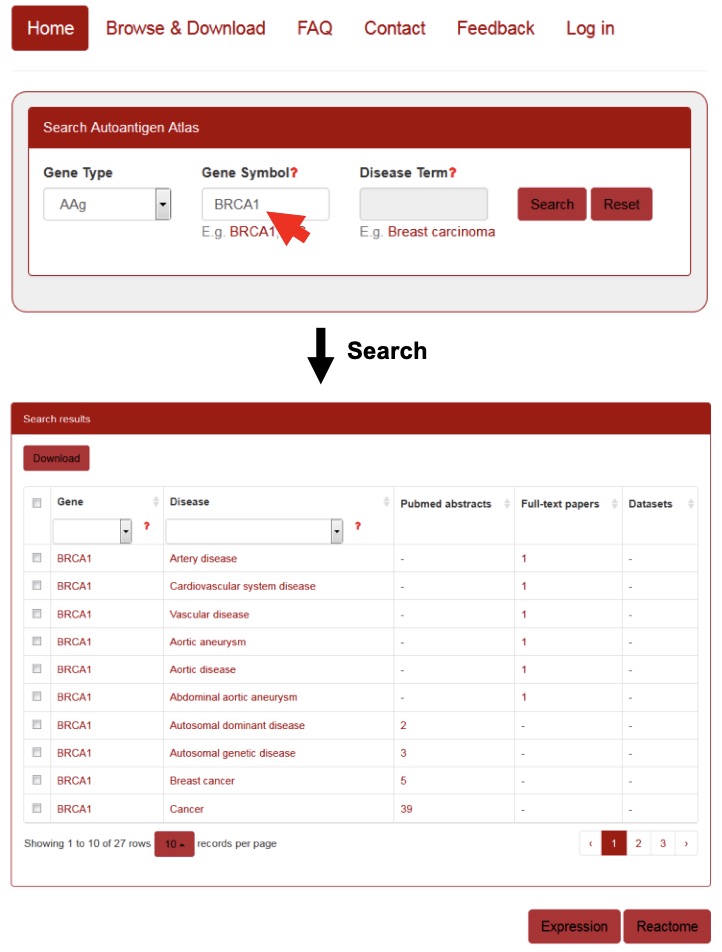
Figure 4. Querying the gene of human AAg in AAg Atlas portal.
For the query by disease term, the user can enter a disease term in the “Disease Term” search box, and a drop-down menu will provide auto-completed disease terms present in the AAgAtlas. After selecting one of them and clicking the ‘Search’ button, the search engine will run and return the queried results containing a table showing the disease term, related AAg genes as well as the supporting evidence. If user click the ‘Reset’ button, all current search terms will be deleted (Figure 5).
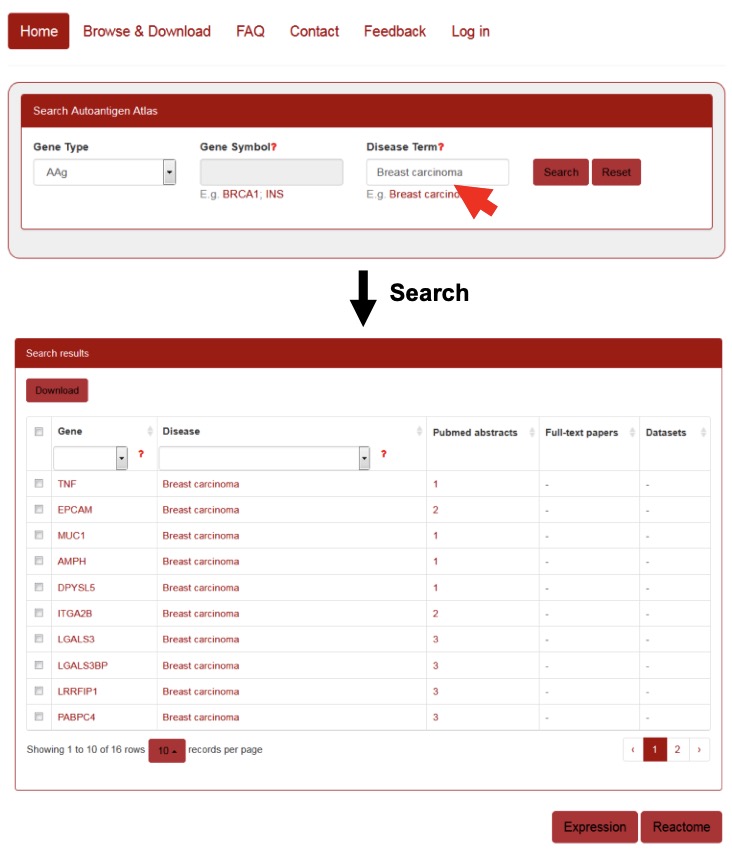
Figure 5. Querying the disease related AAgs in AAgAtlas portal.
By clicking the hyperlink of the gene symbol for an individual AAg in the ‘Gene’ column, users can see the basic information of the AAg gene and the cross references to external databases (i.e. Ensembl, NCBI Gene, UniProtKB, neXtProt and Antibodypedia) (Figure 6).
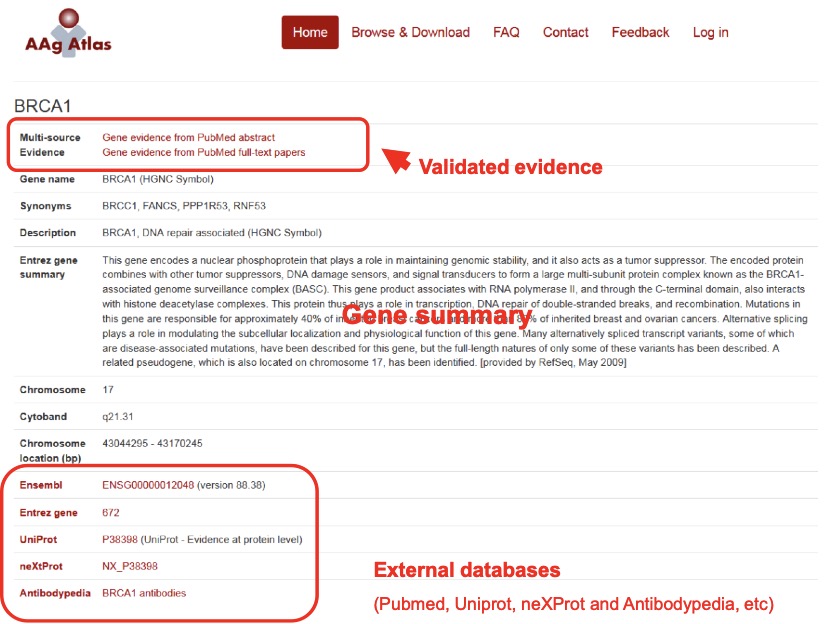
Figure 6. The representative information of the AAg gene of interest.
When the user clicks the small triangle in the head of each column in the table, the correspondent results in the table will be resorted in ascending/descending order by the column. And the user can also input words or select a term in the drop list in the text box to filter the listed diseases from the results by sub-string match. All diseases are accord to Disease Ontology (DO) classification (http://www.disease-ontology.org/) and the detail of each disease is shown after selection (Figure 7).
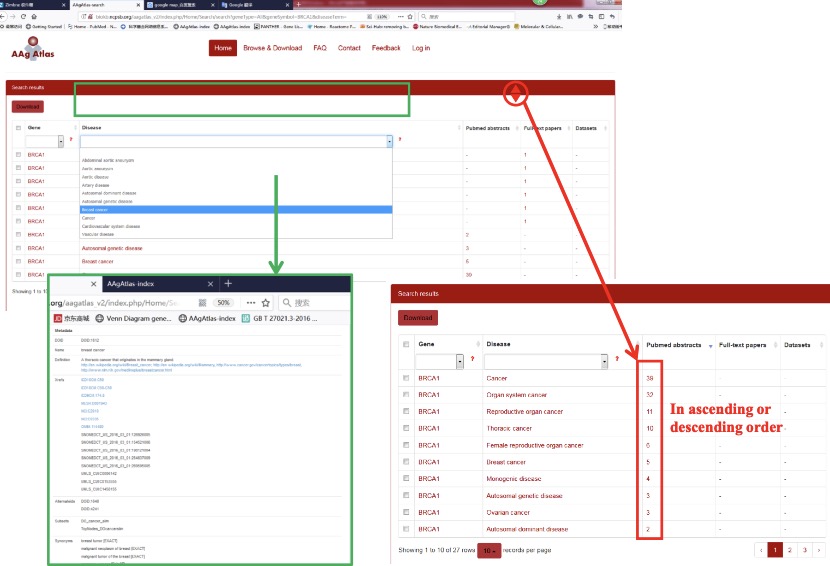
Figure 7. Searching for the information of AAg related human disease.
When the user clicks the number of the evidence abstracts, a table containing gene, disease, the PubMed ID, the evidence sentence and the manual validation information will be displayed. The user can click on the evidence from three resources, including PubMed abstracts, full-text articles and datasets, which are shown below (Figure 8-10).
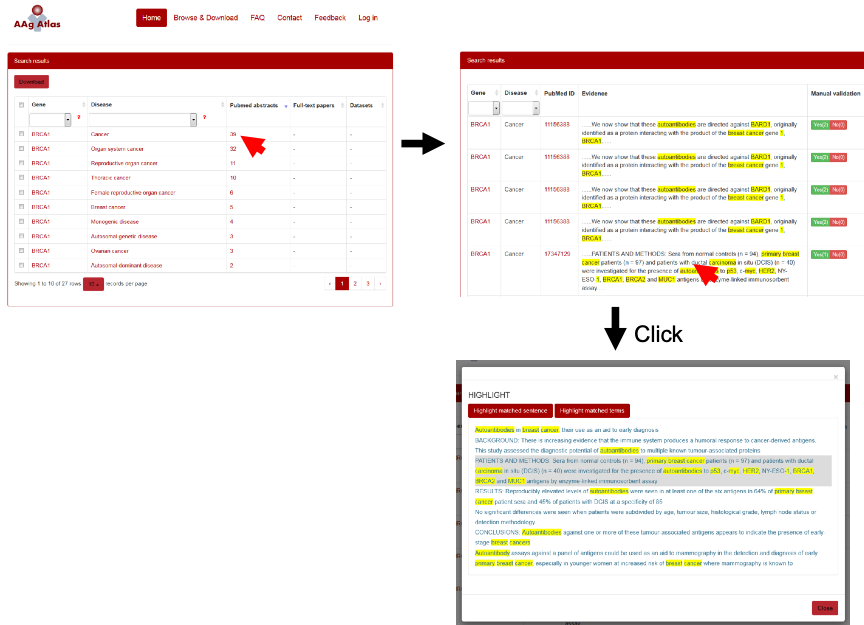
Figure 8. The supporting evidence from PubMed abstracts for human AAgs.
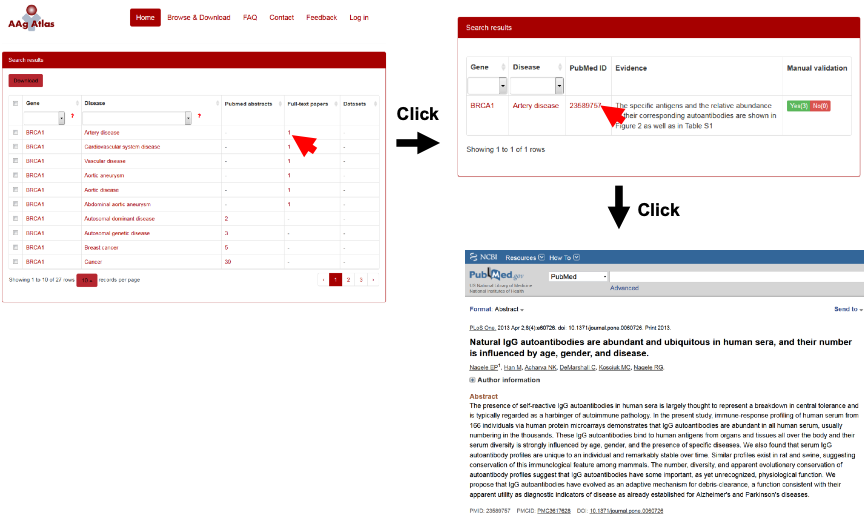
Figure 9. The supporting evidence from PubMed full-text papers for human AAgs.
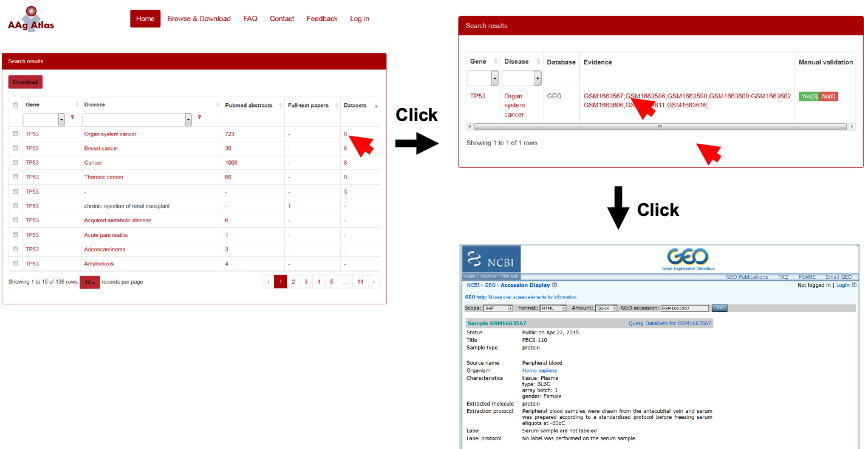
Figure 10. The supporting evidence from microarray databases for human AAgs.
4、How to browse and download the data of AAg or AAg-related diseases?
The list of human AAgs or AAg-related diseases can be browsed by clicking the ‘Browse’ button in the navigation bar. All the information for AAg genes and their supporting literature evidence can be downloaded (red arrow) for further analysis (Figure 11).
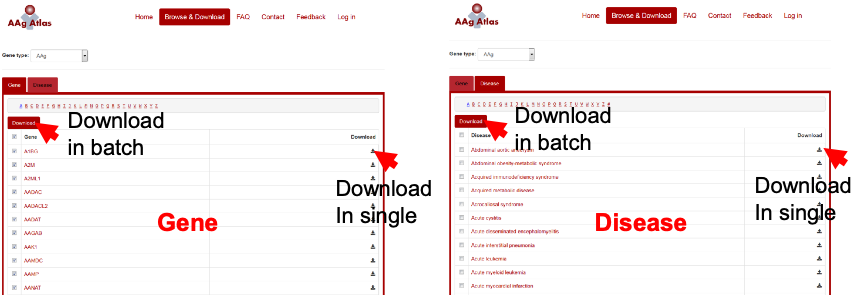
Figure 11. Download of the gene and disease information from AAg Atlas portal.
5、How did our team collect and curate the AAgs and supporting evidence to construct AAg Atlas portal?
Our data collection relies on text mining in AAg-related PubMed abstracts with subsequent manual validation. Bio-entity recognition and extraction from these abstracts for human AAg candidates were performed by a custom ontology-based bio-entity recognizer. After the evaluation against the CRAFT corpus for gene/protein recognition based on the Protein Ontology (PR), the precision, recall, F-measure of our recognition tool are 0.810, 0.883, 0.845, which are on par with current state-of-the-art biomedical annotation systems like BeCAS [Nunes T, et al. Bioinformatics, 2013, 29(15): 1915.].
1) AAgs from PubMed abstracts
We compiled a list of human AAgs together with their related diseases and evidence from PubMed abstract using the following three steps. First, all PubMed abstract were fetched through the NCBI E-utilities API and the AAg-related abstracts were extracted by our custom bio-entity recognizer with the keywords ‘AAg’ or ‘autoantibody’ or their lexical variants like ‘auto-antigen’, ‘auto antigen’, ‘AAgs’, ‘auto-antigens’, ‘auto antigens’, ‘auto-antibody’, ‘auto antibody’, ’autoantibodies’, ‘auto-antibodies’ or ‘autoantibodies’. 45,830 AAg-related abstracts and 94,313 sentences were obtained. Second, a dictionary of human gene/protein name and synonyms was built by integrating all the HGNC-mapped terms and their synonyms in Protein Ontology. A list of human genes was recognized and extracted from the sentences by our bio-entity recognizer based on this dictionary. Gene symbols which co-occurred with the AAg keywords at single-sentence level were considered as candidate AAgs. Third, all 3,984 candidate genes from 25,520 original abstracts and 43,253 evidence sentences in which genes and the keyword co-occurred were manually curated by five experienced researchers for three rounds. Finally, we confirmed 1,126 genes and 1,072 related human diseases, which were used to construct AAgAtlas 1.0 database (http://biokb.ncpsb.org/aagatlas/). The detail is shown in our previous publication in Nucleic Acid Res (Nucleic Acids Res. 2017 45(D1): D769-D776.).
In this work, we greatly expanded AAgAtlas 1.0 database by including AAgs associated with post-translational modification (PTM) and AAgs identified from 1,018 full-text articles as well as in 227 microarray databases (OmicsDI, GEO, AAgMarker, ArrayExpress and PMD) by statistical analysis, which are shown in Figure 1.
2) Selection of post-translational modification related AAgs from PubMed abstracts
We collected AAgs using three approaches (Figure 1). First, we searched PubMed database using “AAg”, “autoantibody” and “post-translational modification” as keywords through text-mining and manual curation as previously described [Nucleic Acids Res. 2017 45(D1): D769-D776.]. The total of 26 PTM related AAgs was identified, including citrullination, acetylation, methylation, glycosylation, phosphorylation, carbamylation, deamidation, hydroxylation and oxidation.
3) AAgs and post-translational modification related AAgs from PubMed papers in full-text and supplementary information.
We searched the Pubmed database using “AAg” or “Autoantibody” and the keywords of proteomics technologies including protein microarray, peptide microarray and mass spectrometry, etc. 1,018 papers were selected in which the full texts and correspondent supplementary documents of 980(96.7%) papers were downloaded. We then manually reviewed those files and identified 6,227 human AAgs and 43 PTM related AAgs (Figure 12).
4) AAgs from microarray databases
Third, we searched OmicsDI, GEO, Array Express, AAgMarker and PMD databases for AAb or AAg screening datasets using protein microarrays. The total of 227 datasets were identified within which 127 datasets using human sera or plasma samples were extracted. After removing redundancy, 99 datasets were selected in which 49 datasets were successfully downloaded. We then executed data analysis in which 13,895 candidates were shown with Z-score higher than 3 in more than two samples. Among of them, 5,485 genes were identified.
Finally, we integrated newly identified AAgs to AAgAtlas 1.0 database and developed next-generation AAg Atlas portal, including 8,382 AAgs and 52 PTM related AAgs and 1,090 related human diseases. All AAg genes/proteins, human disease terms as well as supporting evidence were highlighted and loaded into MySQL database. Moreover, AAg Atlas portal which offers users the interface to query the AAg gene and disease as well as to analyze the expression and biological pathway of AAg genes through Expression Atlas and Reactome via http://biokb.ncpsb.org/aagatlas_V2/index.php. The comparison of AAg Atlas portal and other AAg databases is shown in Figure 3.
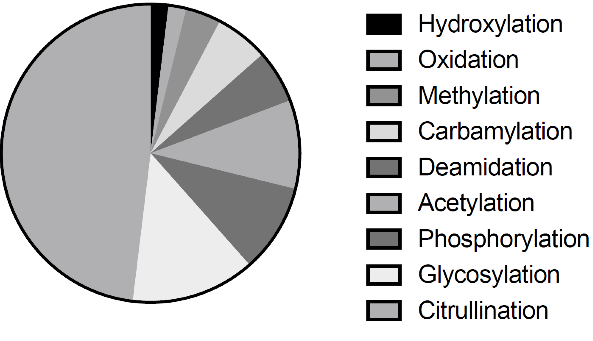
Figure 12. Post-translational modification related AAgs selected in AAg Atlas portal.
6、How the full-text articles were selected?
The full-text articles were selected from PubMed dataset using text mining with “Autoantigen” or “Autoantibody” and the keywords of proteomics technologies as shown in Figure 13.
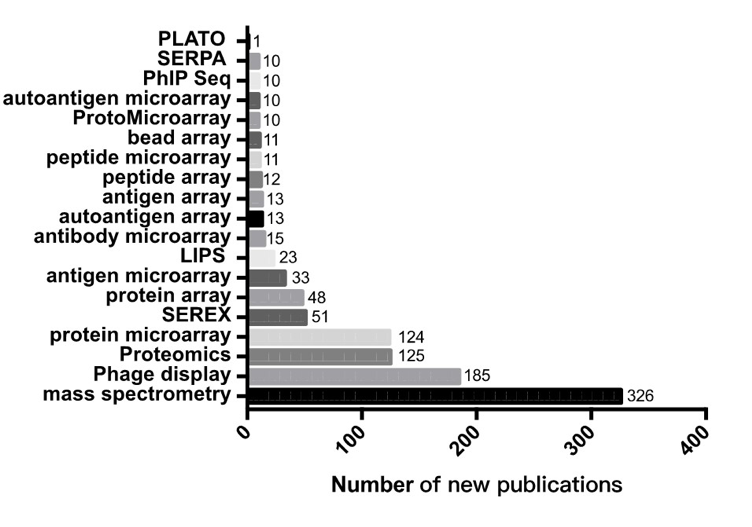
Figure 13. The proteomics technologies used for the identification of human autoantigens from the serum or plasma. SERES is the abbreviation of Serological Analysis of Recombinant cDNA Expression Libraries; LIPS is the abbreviation of Luciferase Immunoprecipitation Systems; SERPA is the abbreviation of Serological Proteome Analysis; PLATO is the abbreviation of Parallel Analysis of in vitro Translational ORFs.
7、How to guarantee data quality in AAg Atlas portal database?
To address this issue, we have taken three rounds of strict manual curation for the AAgs. First, all sentences with AAg names were checked and selected by two experienced researchers independently; second, these selected sentences were submitted to an internal review, in which all AAg names were manually reviewed and approved one by one by a reviewer panel consisting of three experts; third, we asked all co-authors to randomly check AAgs from our website make sure that all genes loaded into our database are bona fide AAgs. Finally, we obtained 8,045 genes and 1,090 related diseases. All the evidence sentences that have been manually curated are regarded as validated evidence and included on the top of each AAg page. In addition, we added a manual curation function to the evidence phrases, with which the users can provide their feedback by simply clicking the “Yes” or “No” button to confirm or reject the evidence phrases. BRCA1 is used as an example and shown in Figure 14.
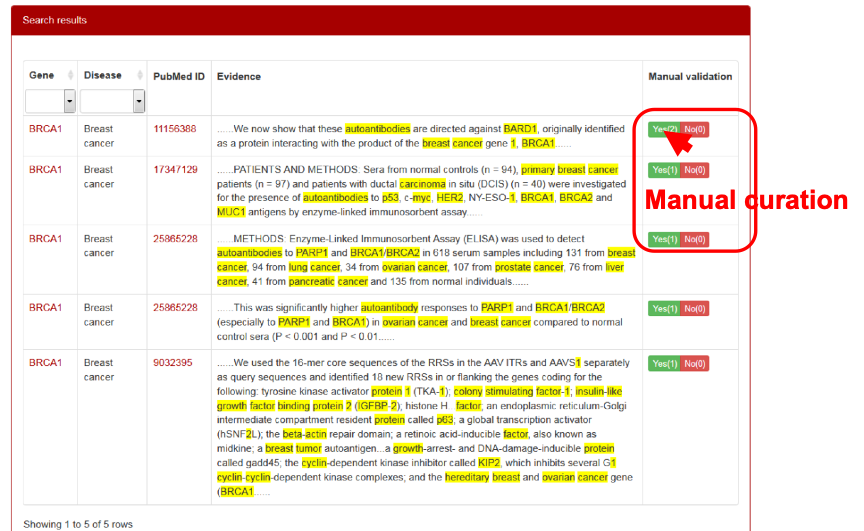
Figure 14. Manual curation of the supporting evidence on AAgAtlas website.
8、How to analyze the expression of target AAg genes through Expression Atlas?
To perform expression analysis, the users can use the genes of AAgs and then submit them by clicking the “Expression” at the bottom. The results will be shown in a new window in which the expression of target genes from different organs will be displayed (Figure 15).
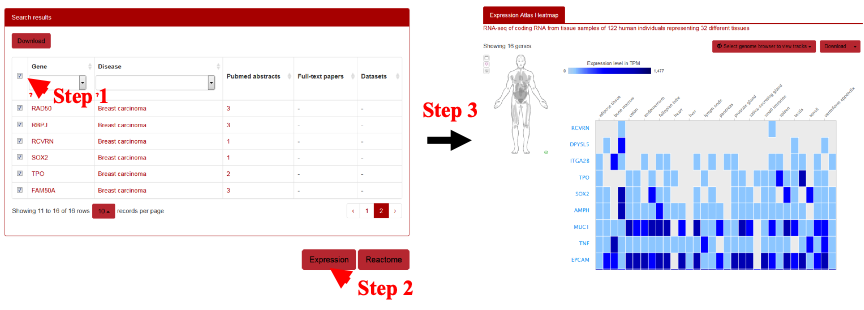
Figure 15. Schematic illustration of gene expression analysis for human autoantigens by Expression Atlas.
9、How to analyze the signaling pathways of target AAg genes through Reactome?
To perform signaling pathway analysis, the users can use the AAg genes of interests and then submit them by clicking the “Reactome” at the right bottom. The results will be shown in a new window in which the signaling pathways of target genes participating will be displayed (Figure 16).
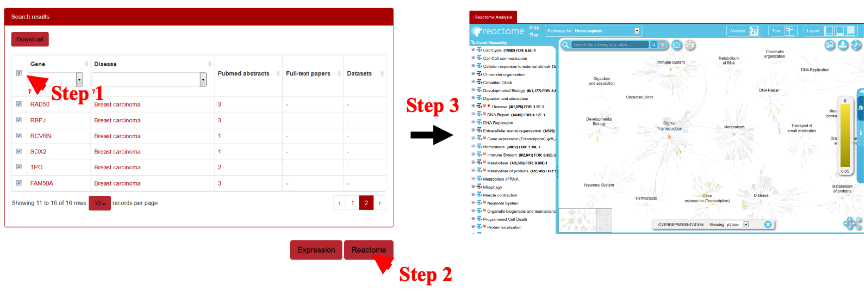
Figure 16. Schematic illustration of signaling pathway analysis for human autoantigens by Reactome.
10、Can the users provide their feedback to our database?
Yes, as described above, we provide a manual annotation function at the end of each evidence phrase, by which the user can confirm the evidence or deny it by simply clicking “Yes” or “No”. We will update our database periodically to include the feedback from the user (Figure 17). The users can also send email to us for further questions or potential collaborations.
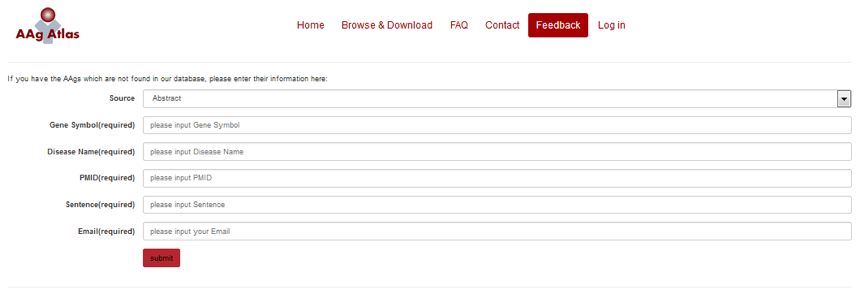
Figure 17. The web location of submitting user’s feedback.
11、Does the user need to log in for manual curation?
Yes, the login is necessary for the users to do the manual curation or download data from our database. We will monitor and validate community curation feedback before inclusion in the database.