1. What is the UVGD?
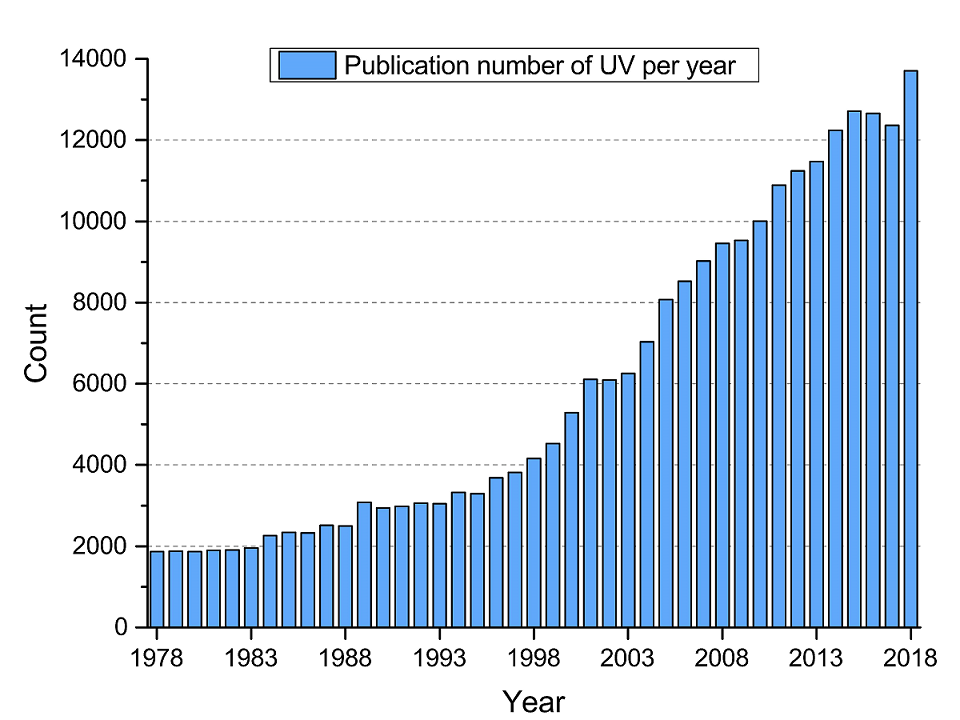
For the past few decades, the number of new publications for ultraviolet radiation is growing rapidly in recent years. However, information about ultraviolet-related genes is dispersed among thousands of publications, and to the best of our knowledge there is no study emphasizing on the systematic collection of ultraviolet-related genes, which brings the obstacle to connect ultraviolet and biological effects in organisms. Under the circumstance, we built UVGD, the UltrViolet-related Gene Database, to bridge ultraviolet radiation and molecular biology effects in organisms. It collected 663 ultraviolet-related genes together with 17 associated biological processes, 117 associated phenotypes and 929 MeSH terms by literature mining and manual curation.
2. How to search the database of UVGD?
Users may search UVGD in four approachs.
First, users can input a gene name in the "Gene Name" search box, or a phenotype name in the "Phenotype" search box, or a biological process name in the "Biological Process" search box, and a drop-down menu will provide auto-completed terms stored in the database. Selecting one of them and clicking the "Search" button will lead to the queried results containing a table showing the ultraviolet-related genes, associated phenotypes, associated biological processes as well as the supporting evidence. All current query terms will be deleted if users click the "Reset" button.
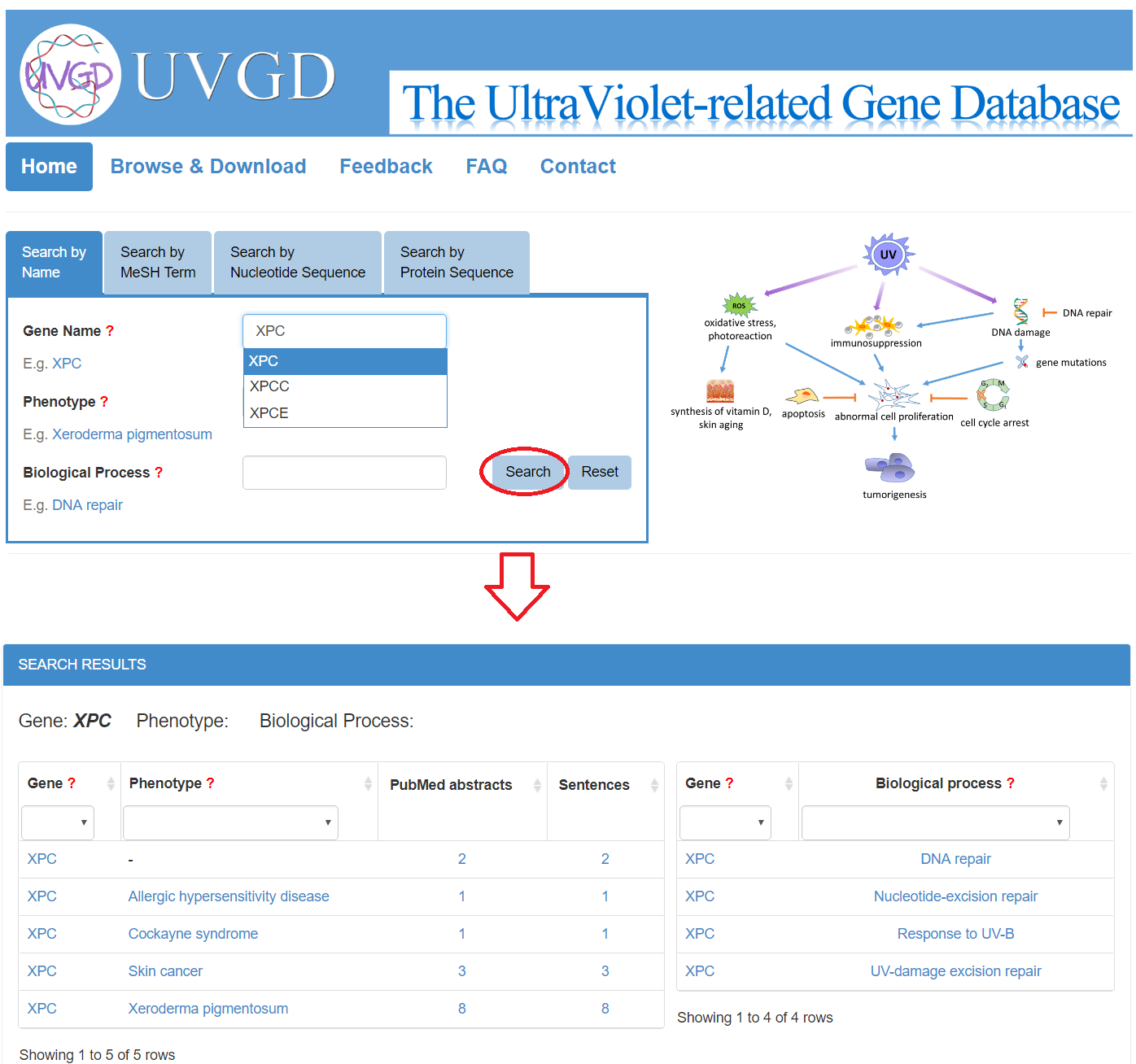
Second, similarly, users can input a MeSH term in the "MeSH Term" search box. By this way, users will get the genes that might be associated with the MeSH term.

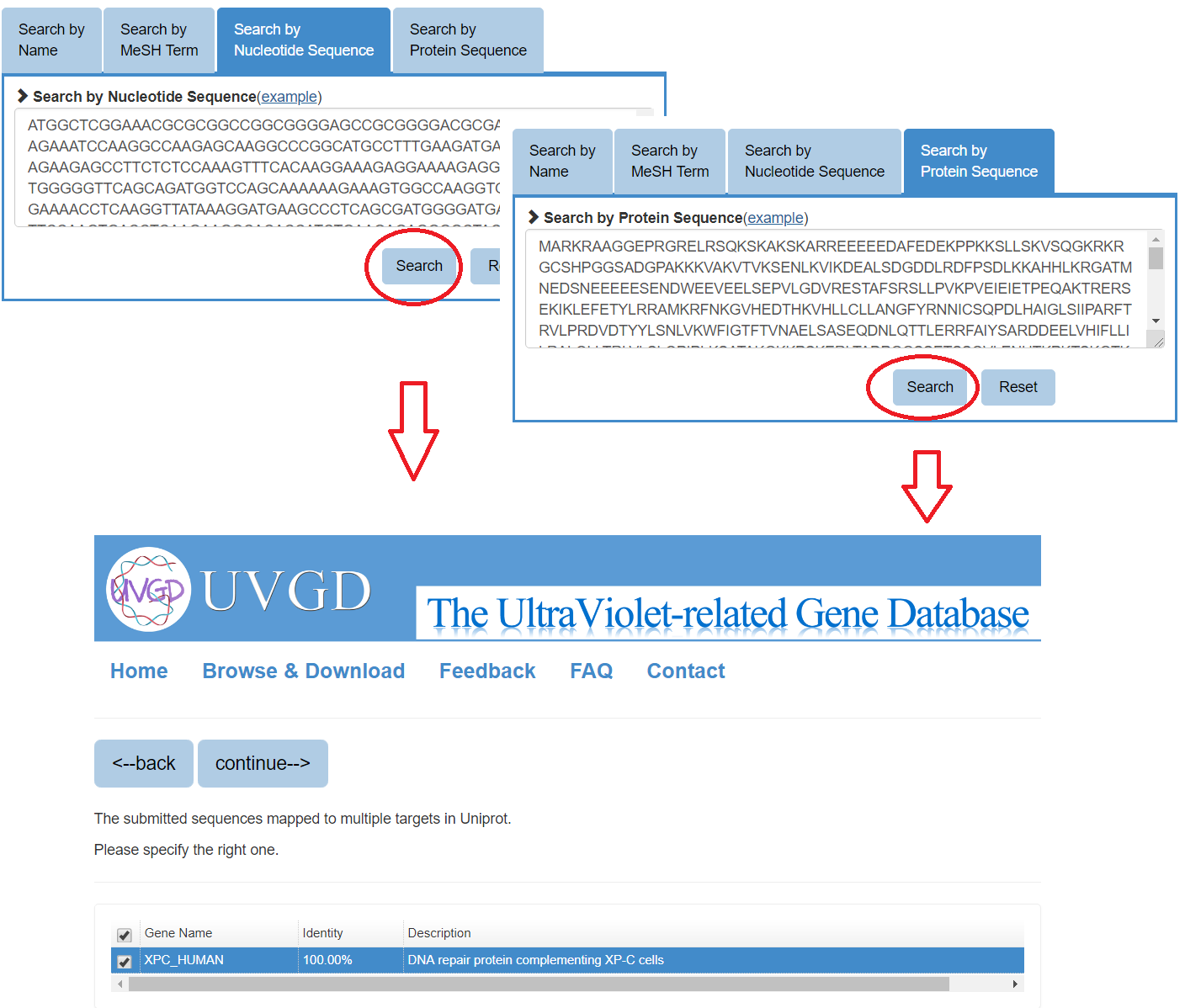
Besides, users also can search the gene by Nucleotide Sequence or Protein Sequence as the third and fourth search approaches, the sequence identity score from BLAST will be listed in the parenthesis after the description. Users can specify the matched gene symbol and click "continue" for result page.
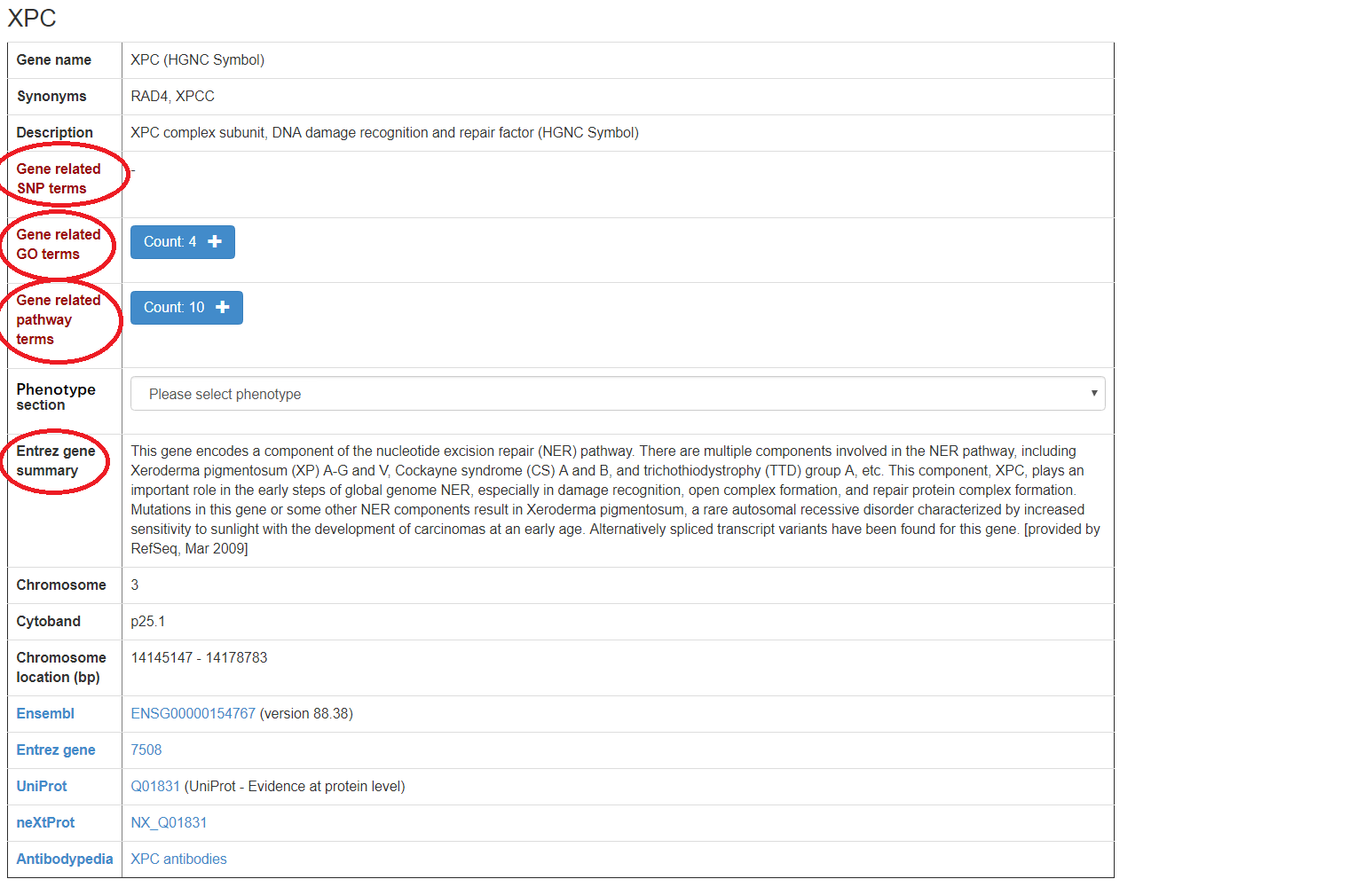
Clicking on the hyperlink of the gene symbol in the "Gene" column will lead to the annotation information of the gene and the cross-references to external databases (i.e. Ensembl, NCBI Gene, UniProtKB, neXtProt, GOC and Antibodypedia).
The user also can input words or select a term in the drop list in the text box to filter the listed genes, phenotypes and biological processes from the results by sub-string match. In addition, clicking the small triangle in the head of each column in the table, the corresponding results in the table will be resorted in ascending/descending order by the column.
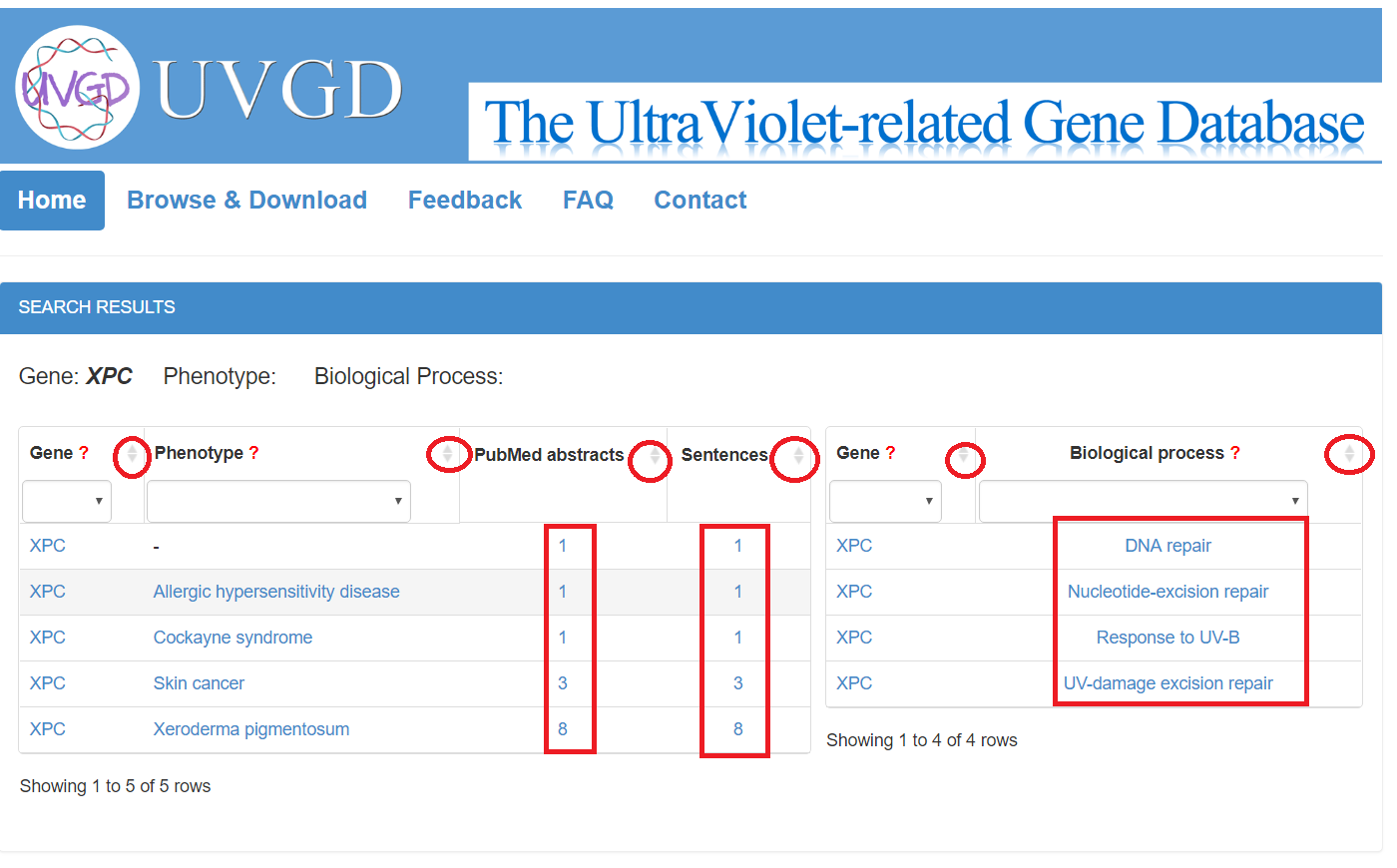
When the user clicks the number of the evidence abstracts or sentences, a table containing gene, phenotype, PubMed ID, organism, the evidence sentence and the manual validation information will be displayed. The user can click on the sentence to see the original abstract in which matched sentence and terms are highlighted in different colours.
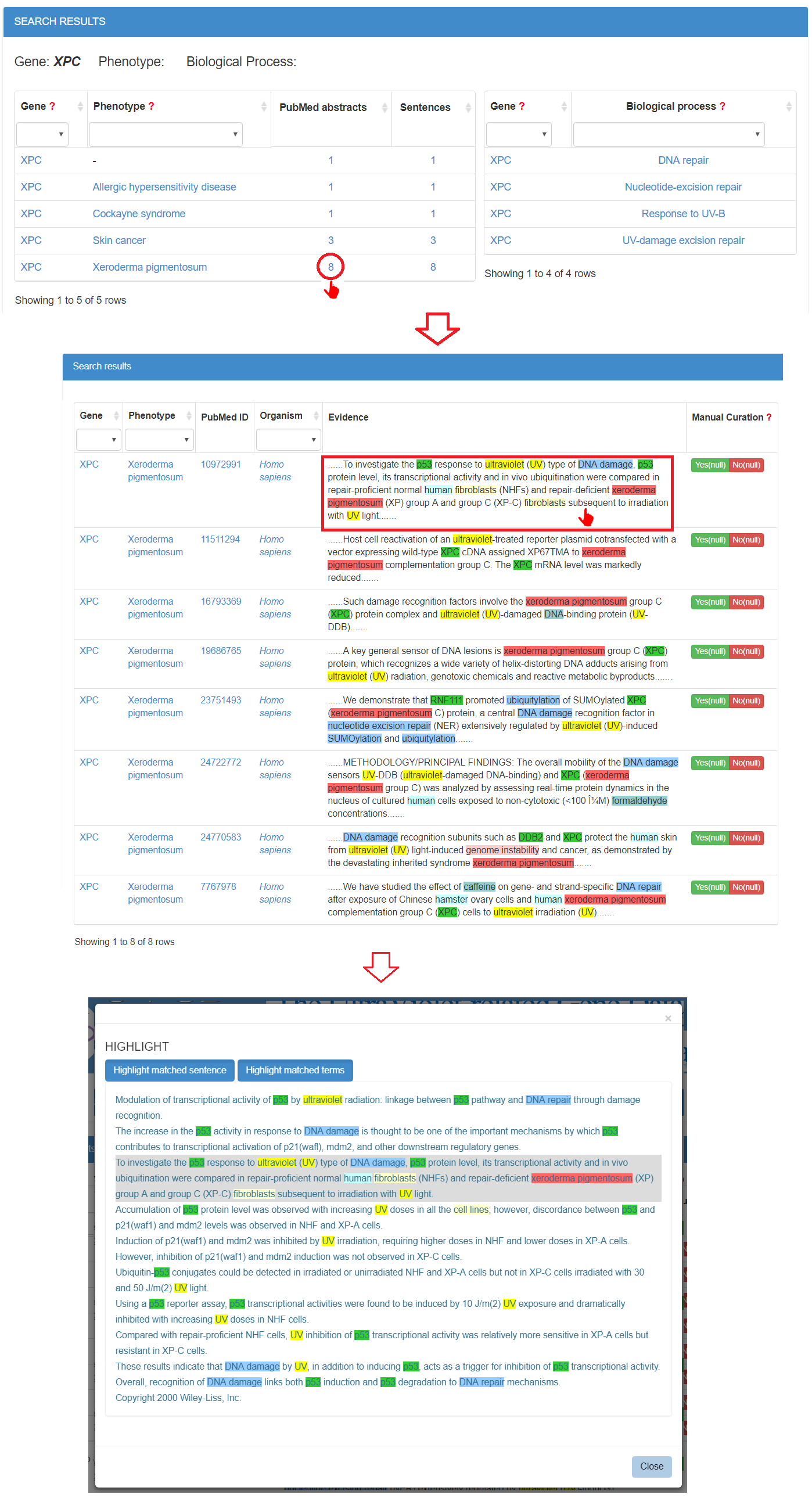
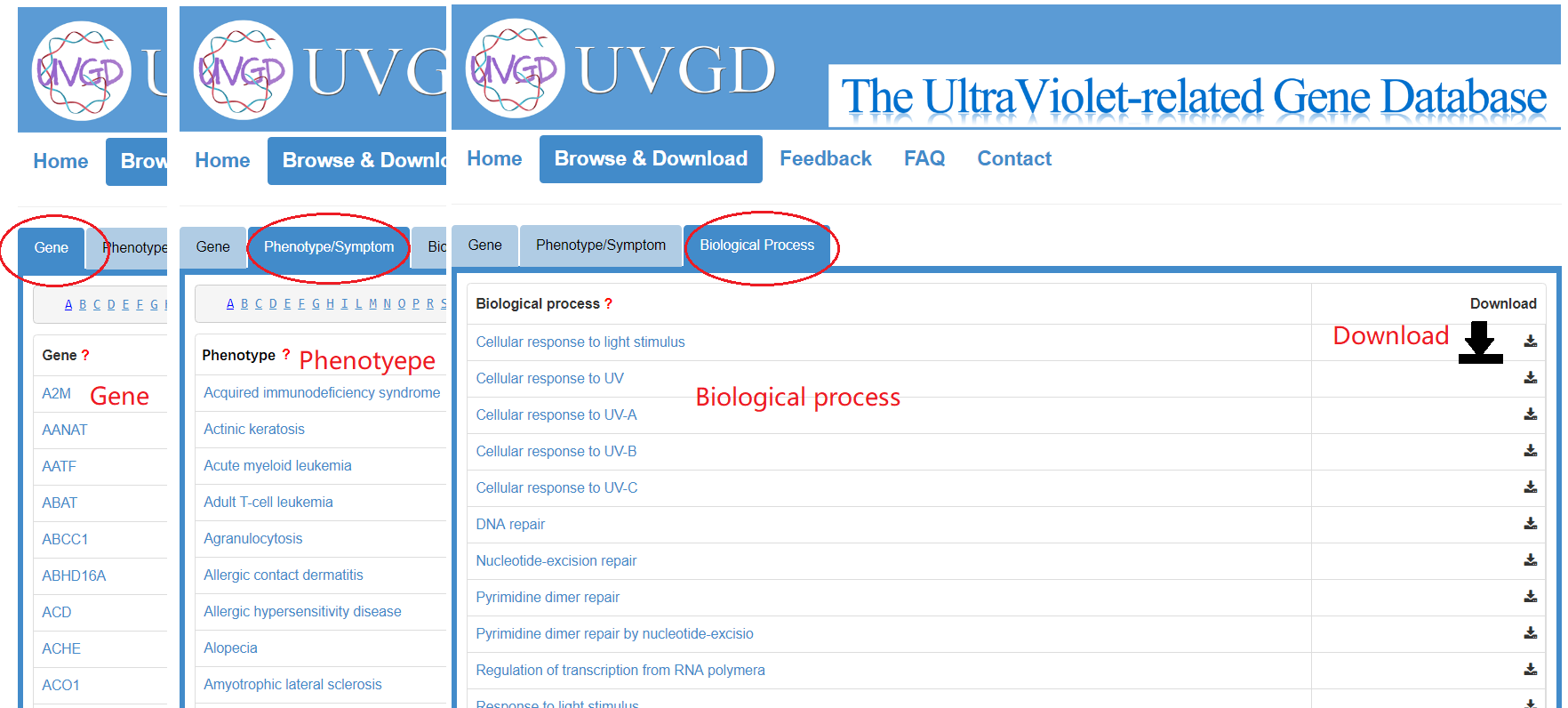
3. How to browse and download data in UVGD?
The complete list of human ultraviolet-related genes and ultraviolet associated phenotypes and biological processes can be browsed by clicking the "Browse & Download" button in the navigation bar. All the information for ultraviolet-related genes and their supporting literature evidence can be downloaded for further analysis.
4. Where are the ultraviolet-related genes, phenotypes, biological processes and MeSH terms from?
We used literature-mining and manual curation to obtain the list of ultraviolet-related genes. These genes with their related evidence was compiled in the following three steps. First, 11,302 PubMed abstracts and 99,471 sentences related to ultraviolet were collected with these keywords: "ultraviolet", "UV", "UVR", "ultra-violet", "ultra violet", "actinic ray", "black light", "antirachitic ray" and their lexical variants. Second, a list of human genes which co-occurred with the keywords related to ultraviolet at single-sentence level were recognized and extracted from these sentences by our bio-entity recognizer. Third, all 2,578 candidate genes with 16,762 sentences were manually curated by our experts, and eventually, 635 genes were finally selected as human ultraviolet-related genes. Besides, we have collected 28 more genes from Gene Ontology Consortium which are described to have solid evidence of being related to ultraviolet.
Phenotype terms collected from Symptom Ontology (SYMP) in EMBL-EBI, while biological process terms collected from Gene Ontology, were also extracted from abstracts based on PubMed. Associations between ultraviolet-related genes and phenotype terms were extracted based on single-sentence level co-occurrence followed by manual curation.
MeSH terms collected from Medical Subject Headings (MeSH). Associations between ultraviolet-related genes and MeSH terms were extracted by processing data from PubMed and MeSH using Python scripts.
5. What is the data quality control in UVGD?
To ensure the data quality, three rounds of strict manual curation were taken for the identification of ultraviolet-related genes. First, all candidate ultraviolet-related genes and supporting evidence were checked by two experienced researchers independently. Second, these selected evidence were submitted to an internal review, in which all ultraviolet-related gene names were manually reviewed one by one by a reviewer team consisting of three experts. Third, all co-authors were asked to randomly check ultraviolet-related genes from our website to make sure that all genes stored in our database are high confidence ultraviolet-related genes. Finally, we obtained 632 genes. All the evidence sentences that have been manually curated are regarded as validated evidence and included on the top of each ultraviolet-related gene page.
6. What is "Manual Curation" by community?
The community can contribute to the manual curation of UVGD by clicking Yes/No in the ‘Manual Curation’ column or input their comments on the evidence phrase by clicking "Comments" button.
To avoid the attack by robot, the login is necessary. Users can log in by clicking here. We will monitor and validate community curation feedback before inclusion in the database.
Here we offer a trial account for the curation function (Account: "test@test.com", Password: "test"). Note: All the other functions of UVGD including data retrieval, browsing and downloading do not require any login or registration.
Note: All the other functions of UVGD including data retrieval, browsing and downloading do not require any login or registration.

7. What is the meaning of "Yes(1) No(0)" in the column of Manual curation?
As it is difficult to validate the massive sentences of co-occurrence between the ultraviolet-related genes and human phenotype terms, we leave the part of manual validation to all logged in users. We added a manual curation function to the sentences, with which the users can provide their feedback by simply clicking the "Yes" or "No" button to confirm or reject the evidence phrases. "Yes (1)"stands for the number of the sentence considered as "right" while "No (0)"stands for the number of the sentence considered as "error".
8. What does "Gene Name" mean?
Gene names mean formal gene symbols and their synonyms provided by RefSeq.
9. What does "Phenotype Term" mean?
Phenotype terms mean standardized phenotype names and their common synonyms provided by Symptom Ontology (SYMP) in EMBL-EBI.
10. What does "Biological Processes" mean?
Biological processes mean standardized biological process names and their common synonyms provided by Gene Ontology.
11. What does "MeSH Term" mean?
MeSH term mean term collected from Medical Subject Headings (MeSH), which covers a wide range of biomedical information, including anatomy; organisms; diseases; chemicals and drugs; analytical, diagnostic and therapeutic techniques, and equipment; psychiatry and psychology; phenomena and processes; disciplines and occupations; anthropology, education, sociology, and social phenomena; technology, industry, and agriculture; information science; named groups; health care; and geographicals.
12. Can the users provide their feedback to the evidence of ultraviolet-related genes?
Yes, "Feedback" feature was provided to our website, with which the users can submit new genes to our database manually; the database will be updated periodically in future. we also added a manual curation function to the evidence sentences of disease terms, with which the users can provide their feedback by simply clicking the "Yes" or "No" button. In addition, the users can also send email to us for further questions or potential collaborations.
